티스토리 뷰
반응형
제1회 신약개발 AI 경진대회
https://dacon.io/competitions/official/236127/overview/description
제1회 신약개발 AI 경진대회 - DACON
분석시각화 대회 코드 공유 게시물은 내용 확인 후 좋아요(투표) 가능합니다.
dacon.io
20여가지 다양한 모델을 대상으로 성능을 평가했으며, 이중에서 성능이 우수한 모델을 선정하였습니다.
모델의 하이퍼 파라메터를 찾기위해 hyperopt 라이브러리를 이용하여 최적화 수행 결과를 DB에 저장하셨습니다.
DB에 저장된 결과에서 최적값을 추출하여 학습을 진행하고, 5개 모델의 결과를 다시한번 Voting Regressor를 이용하여 통합 하였습니다.
사용 모델 : RF, GB, LightGBM, HGB, EXTRATREE
데이터 전처리 및 DB 조회 및 저장
datas.py
import json
import sqlite3
import traceback
import numpy as np
import pandas as pd
from rdkit import DataStructs
from rdkit.Chem import PandasTools, AllChem, Draw
from sklearn.feature_selection import VarianceThreshold
from sklearn.preprocessing import PolynomialFeatures
from xgboost import XGBRegressor
from PIL import Image
def preprocessing(vt_threshold=0.05, fi_threshold=0.0, result_type=None):
features = ["AlogP", "Molecular_Weight", "Num_H_Acceptors", "Num_H_Donors", "Num_RotatableBonds", "LogD",
"Molecular_PolarSurfaceArea"]
train = pd.read_csv('./train.csv', index_col="id")
test = pd.read_csv('./test.csv', index_col="id")
# 중복 데이터 제거(대소문자 구분)
train.drop_duplicates(['SMILES'], keep = False, inplace=True)
# remove 100 over
train = train[train['MLM'] <= 100]
train = train[train['HLM'] <= 100]
# 빈값 채우기
train["AlogP"] = np.where(pd.isna(train["AlogP"]), train["LogD"], train["AlogP"])
test["AlogP"] = np.where(pd.isna(test["AlogP"]), test["LogD"], test["AlogP"])
# SMILES에 포함된 분자를 RDKit 분자로 변환하여 Molecule 컬럼에 추가
PandasTools.AddMoleculeColumnToFrame(train, 'SMILES', 'Molecule')
PandasTools.AddMoleculeColumnToFrame(test, 'SMILES', 'Molecule')
# FPs에 flot 형태로 추가
if result_type == "BINIMG":
train["FPs"] = train.Molecule.apply(mol2binImg)
test["FPs"] = test.Molecule.apply(mol2binImg)
elif result_type == "CHEMIMG":
train["FPs"] = train.Molecule.apply(mol2chemImg)
test["FPs"] = test.Molecule.apply(mol2chemImg)
else:
train["FPs"] = train.Molecule.apply(mol2fp)
test["FPs"] = test.Molecule.apply(mol2fp)
for feature in features:
train["FPs"] = train.apply(lambda x: np.append(x['FPs'], x[feature]), axis=1)
test["FPs"] = test.apply(lambda x: np.append(x['FPs'], x[feature]), axis=1)
# 사용할 column만 추출
train = train[['FPs', 'MLM', 'HLM']].copy()
test = test[['FPs']].copy()
# 특징 선택
transform = VarianceThreshold(vt_threshold)
train_trans = transform.fit_transform(np.stack(train['FPs']))
test_trans = transform.transform(np.stack(test['FPs']))
print("vt_threshold :", vt_threshold, ", feature size =", len(train_trans[0]))
if False:
transform = PolynomialFeatures(degree=2, include_bias=False)
train_trans = transform.fit_transform(train_trans, train['MLM'].values)
test_trans = transform.transform(test_trans)
print("polynomial_features : true", ", feature size =", len(train_trans[0]))
if fi_threshold > 0:
df_fi = feature_importances(train, train_trans)
idx = df_fi[df_fi["fi"] > fi_threshold].index
train_trans = train_trans[:, idx]
test_trans = test_trans[:, idx]
print("fi_threshold :", fi_threshold, ", feature size =", len(train_trans[0]))
train["FPs"] = train_trans.tolist()
test["FPs"] = test_trans.tolist()
return train, test
def mol2fp(mol):
fp = AllChem.GetHashedMorganFingerprint(mol, 6, nBits=4096)
ar = np.zeros((1,), dtype=np.int8)
DataStructs.ConvertToNumpyArray(fp, ar)
return ar
def mol2binImg(mol):
fp = AllChem.GetHashedMorganFingerprint(mol, 6, nBits=4096)
ar = np.zeros((1,), dtype=np.int8)
DataStructs.ConvertToNumpyArray(fp, ar)
img = Image.fromarray((ar * 255).reshape(64, 64))
return np.array(img)
def mol2chemImg(mol):
# ar = Draw.MolToImage(mol, (64, 64))
ar = Draw.MolToImage(mol, (380, 180))
return np.array(ar)
def feature_importances(train_org, trans):
seed_num = 42
# train,valid split
y_mlm = train_org['MLM'].values
y_hlm = train_org['HLM'].values
xgb = XGBRegressor(n_estimators=100)
xgb.fit(trans, y_mlm)
mlm_fi = xgb.feature_importances_
xgb = XGBRegressor(n_estimators=100)
xgb.fit(trans, y_hlm)
hlm_fi = xgb.feature_importances_
df_fi = pd.DataFrame({'mlm': mlm_fi.ravel(), 'hlm': hlm_fi.ravel()})
df_fi['fi'] = (df_fi['mlm'] + df_fi['hlm']) / 2
return df_fi
def insert_optimazer(model_name, column_name, model_param, loss, seed=42):
try:
conn = sqlite3.connect("./db/identifier.sqlite", isolation_level=None)
c = conn.cursor()
c.execute("INSERT INTO optimazer(create_dttm, model_name, column_name, parameter, rmse, seed) \
VALUES(datetime('now','localtime'), ?, ?, ?, ?, ?)", (model_name, column_name, str(model_param), loss, seed))
except:
print("error insert_optimazer()", traceback.format_exc())
def get_optimazer_param(model_name, column_name, seed=42):
try:
conn = sqlite3.connect("./db/identifier.sqlite", isolation_level=None)
c = conn.cursor()
result = c.execute(f"""SELECT * FROM optimazer a
WHERE model_name = '{model_name}' AND column_name = '{column_name}' AND IFNULL(seed, 42) = {seed}
AND rmse = (SELECT min(rmse) FROM optimazer
WHERE model_name = a.model_name AND column_name = a.column_name AND IFNULL(seed, 42) = {seed})
LIMIT 1""").fetchone()
model_param = json.loads(result[3].replace("'", '"').replace("True", 'true').replace("False", 'false'))
print(model_name, column_name, "best parameter =", model_param)
except:
print("database error", traceback.format_exc())
if model_name == "XGB":
model_param = {'colsample_bytree': 0.7376410711316244, 'eta': 0.0763864838648752, 'eval_metric': 'rmse',
'gamma': 5.212511684162327, 'max_depth': 5, 'min_child_weight': 7.0, 'reg_alpha': 151.0,
'reg_lambda': 0.37597466918371075, 'subsample': 0.8843506362879404}
elif model_name == "RF":
model_param = {'max_depth': 16, 'max_features': 0.0804740958473224, 'min_samples_leaf': 1,
'min_samples_split': 7, 'n_estimators': 26}
elif model_name == "TABNET":
model_param = {'gamma': 1.049729273874145, 'lambda_sparse': 0.3850399789109753, 'n_a': 34, 'n_d': 52,
'n_steps': 7}
elif model_name == "KNN":
model_param = {'algorithm': 'ball_tree', 'leaf_size': 49, 'n_neighbors': 30, 'p': 1, 'weights': 'distance'}
elif model_name == "CNN":
model_param = {'batch_size': 32, 'epochs': 100, 'validation_split': 0.18, 'verbose': 1}
else:
model_param = {}
print(model_name, column_name, "best parameter =", model_param)
return model_param
def get_best_models(topn=7):
conn = sqlite3.connect("./db/identifier.sqlite", isolation_level=None)
c = conn.cursor()
result = c.execute(
"""WITH mlm AS (SELECT * FROM optimazer a WHERE seed IS NOT NULL AND column_name = 'MLM' AND IFNULL(seed, 42) != 42 AND (seed, rmse) =
(SELECT seed, MIN(rmse) FROM optimazer WHERE seed = a.seed AND model_name = a.model_name AND column_name = a.column_name LIMIT 1)),
hlm AS (SELECT * FROM optimazer a WHERE seed IS NOT NULL AND column_name = 'HLM' AND IFNULL(seed, 42) != 42 AND (seed, rmse) =
(SELECT seed, MIN(rmse) FROM optimazer WHERE seed = a.seed AND model_name = a.model_name AND column_name = a.column_name LIMIT 1))
SELECT seed, model_name
FROM (
SELECT seed, model_name, rnk
FROM (SELECT seed, model_name, DENSE_RANK() OVER(order BY avg_rmse) rnk
FROM (SELECT seed, model_name, ROUND(AVG(MIN(rmse)) OVER(partition BY seed, column_name), 3) avg_rmse
FROM mlm t
GROUP BY seed, model_name, column_name
ORDER BY avg_rmse, seed) r) s
WHERE rnk <= ?
UNION
SELECT seed, model_name, rnk
FROM (SELECT seed, model_name, DENSE_RANK() OVER(order BY avg_rmse) rnk
FROM (SELECT seed, model_name, ROUND(AVG(MIN(rmse)) OVER(partition BY seed, column_name), 3) avg_rmse
FROM hlm t
GROUP BY seed, model_name, column_name
ORDER BY avg_rmse, seed) r) s
WHERE rnk <= ?) t
GROUP BY seed, model_name""", (topn, topn)).fetchall()
df = pd.DataFrame(result, columns=['seed', 'model_name'])
df = df.groupby(by='seed').agg({'model_name': list})
print(df)
return df
def get_best_rmse_models():
conn = sqlite3.connect("./db/identifier.sqlite", isolation_level=None)
c = conn.cursor()
result = c.execute(
"""SELECT seed, model_name
FROM optimazer
WHERE seed IN (
SELECT seed
FROM (
SELECT * FROM optimazer WHERE column_name = 'MLM' AND rmse < 29.2
UNION
SELECT * FROM optimazer WHERE column_name = 'HLM' AND rmse < 30.3
)
GROUP BY seed
)
GROUP BY seed, model_name""").fetchall()
df = pd.DataFrame(result, columns=['seed', 'model_name'])
print(df)
return df
모델 학습, 예측, 파라메터 튜닝 관련
models.py
import os
import json
import random
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier, ExtraTreesRegressor
from xgboost import XGBRegressor
from sklearn.ensemble import HistGradientBoostingRegressor
from lightgbm import LGBMRegressor
from torch.utils.data import Dataset, DataLoader
from hyperopt import STATUS_OK, Trials, fmin, hp, tpe, space_eval
from sklearn.metrics import mean_squared_error
from sklearn.metrics import accuracy_score
class BinaryClassification(nn.Module):
def __init__(self, x_train):
super(BinaryClassification, self).__init__()
self.layer_1 = nn.Linear(x_train.shape[1], 64)
self.layer_2 = nn.Linear(64, 64)
self.layer_out = nn.Linear(64, 1)
self.relu = nn.ReLU()
self.dropout = nn.Dropout(p=0.1)
self.batchnorm1 = nn.BatchNorm1d(64)
self.batchnorm2 = nn.BatchNorm1d(64)
self.seed = 42
def forward(self, inputs):
x = self.layer_1(inputs)
x = self.relu(x)
x = self.batchnorm1(x)
x = self.layer_2(x)
x = self.relu(x)
x = self.batchnorm2(x)
x = self.dropout(x)
x = self.layer_out(x)
return x
class CustomDataset(Dataset):
def __init__(self, x_data, y_data=None):
self.x_data = x_data
self.y_data = y_data
def __getitem__(self, index):
if self.y_data is not None:
return self.x_data[index], self.y_data[index]
else:
return self.x_data[index]
def __len__(self):
return len(self.x_data)
class Model_collections:
def __init__(self, model_name):
self.model_name = model_name
self.model = None
self.data = None
def model_fit(self, x_train, y_train, model_param):
model_param = json.loads(model_param) if type(model_param) == str else model_param
if self.model_name == 'XGB':
self.model = self.XGBoost(x_train, y_train, model_param)
return self.model
elif self.model_name == 'RF':
self.model = self.RandomForest(x_train, y_train, model_param)
return self.model
elif self.model_name == "LIGHTGBM":
self.model = self.LightGBM(x_train, y_train, model_param)
return self.model
elif self.model_name == 'HGB':
self.model = self.HGBoost(x_train, y_train, model_param)
return self.model
elif self.model_name == 'EXTRATREE':
self.model = self.ExtraTree(x_train, y_train, model_param)
return self.model
else:
print("Invalid model.")
return None
@staticmethod
def XGBoost(x_train, y_train, model_param):
XGBoost = XGBRegressor(**model_param)
model = XGBoost.fit(x_train, y_train)
return model
@staticmethod
def RandomForest(x_train, y_train, model_param):
RF = RandomForestRegressor(**model_param)
model = RF.fit(x_train, y_train.ravel())
return model
@staticmethod
def LightGBM(x_train, y_train, model_param):
model_param['verbose'] = -1
model = LGBMRegressor(**model_param)
model.fit(x_train, y_train.ravel())
return model
@staticmethod
def HGBoost(x_train, y_train, model_param):
HGBoost = HistGradientBoostingRegressor(**model_param)
model = HGBoost.fit(x_train, y_train.ravel())
return model
@staticmethod
def ExtraTree(x_train, y_train, model_param):
ET = ExtraTreesRegressor(**model_param)
model = ET.fit(x_train, y_train.ravel())
return model
def get_optimazer_space(self, random_state=42):
self.seed = random_state
if self.model_name == 'XGB':
# https://xgboost.readthedocs.io/en/stable/parameter.html
space = {'eta': hp.uniform('eta', 0, 1),
'subsample': hp.uniform('subsample', 0, 1),
'max_depth': hp.uniformint("max_depth", 3, 20),
'gamma': hp.uniform('gamma', 1, 10),
'reg_alpha': hp.quniform('reg_alpha', 60, 180, 1),
'reg_lambda': hp.uniform('reg_lambda', 0, 1),
'colsample_bytree': hp.uniform('colsample_bytree', 0.5, 1),
'min_child_weight': hp.quniform('min_child_weight', 0, 15, 1),
'eval_metric': 'rmse',
'random_state': random_state,
'n_jobs': 20}
return space
elif self.model_name == 'RF':
# https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestRegressor.html
space = {'n_estimators': hp.uniformint('n_estimators', 50, 120),
'max_depth': hp.uniformint('max_depth', 10, 25), # 3 ~ 20
'min_samples_split': hp.uniformint('min_samples_split', 2, 15), # 2 ~ 10
'min_samples_leaf': hp.uniformint('min_samples_leaf', 1, 5),
'max_features': hp.uniform('max_features', 0, 1),
'random_state': random_state,
'n_jobs': 20}
return space
elif self.model_name == "LIGHTGBM":
# https://lightgbm.readthedocs.io/en/stable/Parameters.html
space = {'num_leaves': hp.uniformint('num_leaves', 31, 200),
'tree_learner': hp.choice('tree_learner', ['serial', 'feature', 'data', 'voting']),
'learning_rate': hp.uniform('learning_rate', 0, 0.2),
'max_depth': hp.uniformint('max_depth', 1, 20),
'reg_lambda': hp.uniform('reg_lambda', 0, 1),
'subsample': hp.uniform('subsample', 0, 1),
'random_state': random_state,
'verbose': -1,
'n_jobs': 20}
return space
elif self.model_name == 'HGB':
# https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.HistGradientBoostingRegressor.html
space = {'learning_rate': hp.uniform('learning_rate', 0, 0.2),
'max_iter': hp.uniformint('max_iter', 1, 5) * 100,
'max_leaf_nodes': hp.uniformint('max_leaf_nodes', 2, 50),
'max_depth': hp.uniformint('max_depth', 3, 20),
'min_samples_leaf': hp.uniformint('min_samples_leaf', 1, 10),
'random_state': random_state}
return space
elif self.model_name == 'EXTRATREE':
space = {'n_estimators': hp.uniformint('n_estimators', 200, 800),
'max_depth': hp.uniformint('max_depth', 70, 120), # 3 ~ 20
'min_samples_split': hp.uniformint('min_samples_split', 3, 10), # 2 ~ 10
'min_samples_leaf': hp.uniformint('min_samples_leaf', 1, 2),
'min_weight_fraction_leaf': hp.uniform('min_weight_fraction_leaf', 0, 0.00001),
'max_features': hp.uniform('max_features', 0.1, 0.3),
'random_state': random_state,
'n_jobs': 20}
return space
else:
print("Invalid model.")
return None
@staticmethod
def set_seed(seed=42):
random.seed(seed)
np.random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
os.environ["HYPEROPT_FMIN_SEED"] = str(seed)
torch.manual_seed(seed)
def optimazer(self, data, space=None, debug=False, max_evals=None):
trials = Trials()
self.data = data
if space is None:
space = self.get_optimazer_space()
if space is None:
print("모델의 옵티마이저 파라메터를 설정하지 않았습니다.")
exit(0)
if max_evals is None:
if self.model_name in ['HGB']:
max_evals = 50
elif self.model_name in ['RF', 'XGB', 'LIGHTGBM', 'EXTRATREE']:
max_evals = 100
else:
max_evals = 100
self.set_seed(self.seed)
best_hyperparams = fmin(fn=self.objective, space=space, algo=tpe.suggest, max_evals=max_evals, trials=trials)
loss = trials.best_trial['result']['loss']
best_hyperparams = space_eval(space, best_hyperparams)
df_loss = pd.DataFrame(trials.results)
df_vals = pd.DataFrame(trials.vals)
df_vals['loss'] = df_loss['loss']
df_vals.sort_values(by=['loss'], inplace=True)
if debug:
pd.set_option('display.max_columns', None)
pd.set_option('display.width', 130)
print(df_vals.head(15))
pd.reset_option("display.max_columns")
pd.reset_option("display.width")
if debug:
print(self.model_name, "best loss :", loss, "best hyper parameters :", best_hyperparams)
return loss, best_hyperparams
def objective(self, space):
X_train = self.data['X_train']
y_train = self.data['y_train']
X_test = self.data['X_test']
y_test = self.data['y_test']
clf = self.model_fit(X_train, y_train, space)
pred = clf.predict(X_test)
if self.model_name.endswith("-CF"):
accuracy = accuracy_score(y_test, pred > 0.5)
return {'loss': -accuracy, 'status': STATUS_OK}
else:
rmse = mean_squared_error(y_test, pred, squared=False)
return {'loss': rmse, 'status': STATUS_OK}
하이퍼 파라메터 최적화
main_optimazer.py
import random
import os
import numpy as np
import datetime
from sklearn.model_selection import train_test_split
import datas
from models import *
def set_seed(seed=42):
random.seed(seed)
np.random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
torch.manual_seed(seed)
def validation_output(mlm, hlm, valid_df, model_name, start_tm):
rmse_mlm = mean_squared_error(valid_df['MLM'].values, mlm, squared=False)
rmse_hlm = mean_squared_error(valid_df['HLM'].values, hlm, squared=False)
rmse = round((rmse_mlm + rmse_hlm) / 2, 2)
valid_df['predMLM'] = mlm
valid_df['predHLM'] = hlm
# valid_df.to_csv(f'./output/validation_{model_name}_{start_tm}_rmse{rmse}.csv')
# print(valid_df)
print(start_tm, "model_name =", model_name, " RMSE =", rmse, ", MLM_RMSE =", rmse_mlm, ", HLM_RMSE =", rmse_hlm)
def submission(mlm, hlm, start_tm):
submission = pd.read_csv('submission_format.csv')
submission['MLM'] = mlm
submission['HLM'] = hlm
submission.to_csv(f'./output/submission_{start_tm}.csv', index=False)
if __name__ == '__main__':
# 랜덤 시드
seed_nums = [255]
# seed_nums = range(1, 400)
# 모델
# model_names = ['EXTRATREE']
model_names = ['RF', 'XGB', 'LIGHTGBM', 'HGB', 'EXTRATREE']
# 최대반복횟수(None이면 models에 지정된 값 사용)
# max_evals = 100
max_evals = None
for seed_num in seed_nums:
for model_name in model_names:
print("seed_num = ", seed_num, ", model = ", model_name)
start_tm = datetime.datetime.now().strftime("%Y%m%d_%H%M%S")
set_seed(seed_num)
# preprocessing
if model_name == "CNN":
train, test = datas.preprocessing(vt_threshold=0.05, fi_threshold=0.0, result_type="BINIMG")
else:
train, test = datas.preprocessing(vt_threshold=0.05, fi_threshold=0.0)
if model_name.endswith("-CF"):
train['MLM'] = train.apply(lambda x: 0 if x['MLM'] < 20 else 9 if x['MLM'] > 80 else 5, axis=1)
train['HLM'] = train.apply(lambda x: 0 if x['HLM'] < 20 else 9 if x['HLM'] > 80 else 5, axis=1)
# train,valid split
train_x, valid_x = train_test_split(train, test_size=0.2, random_state=seed_num)
# model create
mc = Model_collections(model_name)
# optimazer
space = mc.get_optimazer_space(random_state=seed_num)
if space is None:
print("모델의 옵티마이저 파라메터를 설정하지 않았습니다.")
exit(0)
data = {"X_train": np.stack(train_x['FPs']), "y_train": train_x['MLM'].values.reshape(-1, 1),
"X_test": np.stack(valid_x['FPs']), "y_test": valid_x['MLM'].values.reshape(-1, 1)}
mlm_loss, model_param_mlm = mc.optimazer(data, space=space, debug=True, max_evals=max_evals)
datas.insert_optimazer(model_name, 'MLM', model_param_mlm, mlm_loss, seed_num)
data = {"X_train": np.stack(train_x['FPs']), "y_train": train_x['HLM'].values.reshape(-1, 1),
"X_test": np.stack(valid_x['FPs']), "y_test": valid_x['HLM'].values.reshape(-1, 1)}
hlm_loss, model_param_hlm = mc.optimazer(data, space=space, debug=True, max_evals=max_evals)
datas.insert_optimazer(model_name, 'HLM', model_param_hlm, hlm_loss, seed_num)
# # training
model_MLM = mc.model_fit(np.stack(train_x['FPs']), train_x['MLM'].values.reshape(-1, 1), model_param_mlm)
model_HLM = mc.model_fit(np.stack(train_x['FPs']), train_x['HLM'].values.reshape(-1, 1), model_param_hlm)
# validation
vpred_MLM = model_MLM.predict(np.stack(valid_x['FPs']))
vpred_HLM = model_HLM.predict(np.stack(valid_x['FPs']))
validation_output(vpred_MLM, vpred_HLM, valid_x[['MLM', 'HLM']], model_name, start_tm)
print()
# # predict
# pred_MLM = model_MLM.predict(np.stack(test['FPs']))
# pred_HLM = model_HLM.predict(np.stack(test['FPs']))
#
# submission(pred_MLM, pred_HLM, start_tm)
학습 및 예측
main_voting.py
import datetime
from sklearn.model_selection import train_test_split
from sklearn.ensemble import VotingRegressor
from sklearn.preprocessing import MinMaxScaler
import matplotlib.pyplot as plt
import datas
from models import *
def set_seed(seed=42):
random.seed(seed)
np.random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
torch.manual_seed(seed)
def validation_output(mlm, hlm, valid_df, model_name, start_tm):
rmse_mlm = mean_squared_error(valid_df['MLM'].values, mlm, squared=False)
rmse_hlm = mean_squared_error(valid_df['HLM'].values, hlm, squared=False)
rmse = round((rmse_mlm + rmse_hlm) / 2, 2)
print(start_tm, "model_name =", model_name, " RMSE =", rmse, ", MLM_RMSE =", rmse_mlm, ", HLM_RMSE =", rmse_hlm)
return rmse_mlm, rmse_hlm
def submission(mlm, hlm, start_tm):
submission = pd.read_csv('submission_format.csv')
submission['MLM'] = mlm
submission['HLM'] = hlm
submission.to_csv(f'./output/submission_{start_tm}.csv', index=False)
def plot_voting(pred_mdl, pred_ereg, us_pred_ereg, real):
plt.figure(figsize=(20, 10))
for i, pred in enumerate(pred_mdl):
if i == 0:
plt.plot(pred[1][:20], "yd", label=pred[0])
elif i == 1:
plt.plot(pred[1][:20], "gd", label=pred[0])
elif i == 2:
plt.plot(pred[1][:20], "bd", label=pred[0])
else:
plt.plot(pred[1][:20], "gd", label=pred[0])
plt.plot(pred_ereg[:20], "b*", ms=10, label="VOTING")
plt.plot(us_pred_ereg[:20], "r*", ms=10, label="VOTING-US")
plt.plot(real[:20], "ro", ms=10, label="REAL")
plt.tick_params(axis="x", which="both", bottom=False, top=False, labelbottom=False)
plt.ylabel("predicted")
plt.xlabel("training samples")
plt.legend(loc="best")
plt.title("Regressor predictions and their average")
plt.show()
if __name__ == '__main__':
seed_num = 396
start_tm = datetime.datetime.now().strftime("%Y%m%d_%H%M%S")
model_names = ['RF', 'XGB', 'LIGHTGBM', 'HGB', 'EXTRATREE']
# preprocessing
set_seed(seed_num)
train, test = datas.preprocessing()
# train,valid split
train_x, valid_x = train_test_split(train, test_size=0.2, random_state=seed_num)
mlm_models, hlm_models = [], []
# fit and predict and performance
for model_name in model_names:
set_seed(seed_num)
mc = Model_collections(model_name)
model_mlm = mc.model_fit(np.stack(train_x['FPs']), train_x['MLM'].values.reshape(-1, 1),
datas.get_optimazer_param(model_name, "MLM", seed_num))
model_hlm = mc.model_fit(np.stack(train_x['FPs']), train_x['HLM'].values.reshape(-1, 1),
datas.get_optimazer_param(model_name, "HLM", seed_num))
pred_mlm = model_mlm.predict(np.stack(valid_x['FPs']))
pred_hlm = model_hlm.predict(np.stack(valid_x['FPs']))
rmse_mlm, rmse_hlm = validation_output(pred_mlm, pred_hlm, valid_x[['MLM', 'HLM']], model_name, start_tm)
mlm_models.append((model_name, model_mlm, rmse_mlm, pred_mlm))
hlm_models.append((model_name, model_mlm, rmse_hlm, pred_hlm))
# voting model choose
mlm_models = sorted(mlm_models, key=lambda x: x[2])
hlm_models = sorted(hlm_models, key=lambda x: x[2])
voting_model_mlm, voting_model_hlm = [], []
plot_pred_mlm, plot_pred_hlm = [], []
use_models = 4
print("Best models:")
for i in range(len(mlm_models)):
if i < use_models:
print("Rank", i + 1, "* : MLM =", mlm_models[i][0], mlm_models[i][2], "HLM =", hlm_models[i][0],
hlm_models[i][2])
voting_model_mlm.append((mlm_models[i][0], mlm_models[i][1]))
voting_model_hlm.append((hlm_models[i][0], hlm_models[i][1]))
plot_pred_mlm.append((mlm_models[i][0], mlm_models[i][3]))
plot_pred_hlm.append((hlm_models[i][0], hlm_models[i][3]))
else:
print("Rank", i + 1, " : MLM =", mlm_models[i][0], mlm_models[i][2], "HLM =", hlm_models[i][0],
hlm_models[i][2])
# voting and valid predict
ereg_mlm = VotingRegressor(voting_model_mlm)
ereg_hlm = VotingRegressor(voting_model_hlm)
ereg_mlm.fit(np.stack(train_x['FPs']), train_x['MLM'].values.reshape(-1, 1).ravel())
ereg_hlm.fit(np.stack(train_x['FPs']), train_x['HLM'].values.reshape(-1, 1).ravel())
pred_ereg_mlm = ereg_mlm.predict(np.stack(valid_x['FPs']))
pred_ereg_hlm = ereg_hlm.predict(np.stack(valid_x['FPs']))
validation_output(pred_ereg_mlm, pred_ereg_hlm, valid_x[['MLM', 'HLM']], "VOTING", start_tm)
# inverse scale
scaler = MinMaxScaler(feature_range=(np.min(pred_ereg_mlm), np.max(pred_ereg_mlm)))
scaler.fit(train_x['MLM'].values.reshape(-1, 1)) ## 각 칼럼 데이터마다 변환할 함수 생성
us_pred_ereg_mlm = scaler.inverse_transform(pred_ereg_mlm.reshape(-1, 1)) ## 원 데이터의 스케일로 변환
scaler = MinMaxScaler(feature_range=(np.min(pred_ereg_hlm), np.max(pred_ereg_hlm)))
scaler.fit(train_x['HLM'].values.reshape(-1, 1)) ## 각 칼럼 데이터마다 변환할 함수 생성
us_pred_ereg_hlm = scaler.inverse_transform(pred_ereg_hlm.reshape(-1, 1)) ## 원 데이터의 스케일로 변환
rmse_mlm, rmse_hlm = validation_output(us_pred_ereg_mlm, us_pred_ereg_hlm, valid_x[['MLM', 'HLM']],
"VOTING UP-SCALE", start_tm)
plot_voting(plot_pred_mlm, pred_ereg_mlm, us_pred_ereg_mlm, valid_x['MLM'].values)
plot_voting(plot_pred_hlm, pred_ereg_hlm, us_pred_ereg_hlm, valid_x['HLM'].values)
# ==================================================================================================================
# test predict
pred_ereg_mlm = ereg_mlm.predict(np.stack(test['FPs']))
pred_ereg_hlm = ereg_hlm.predict(np.stack(test['FPs']))
# inverse scale
scaler = MinMaxScaler(feature_range=(np.min(pred_ereg_mlm), np.max(pred_ereg_mlm)))
scaler.fit(train_x['MLM'].values.reshape(-1, 1)) ## 각 칼럼 데이터마다 변환할 함수 생성
pred_ereg_mlm = scaler.inverse_transform(pred_ereg_mlm.reshape(-1, 1)) ## 원 데이터의 스케일로 변환
scaler = MinMaxScaler(feature_range=(np.min(pred_ereg_hlm), np.max(pred_ereg_hlm)))
scaler.fit(train_x['HLM'].values.reshape(-1, 1)) ## 각 칼럼 데이터마다 변환할 함수 생성
pred_ereg_hlm = scaler.inverse_transform(pred_ereg_hlm.reshape(-1, 1)) ## 원 데이터의 스케일로 변환
# ensemble
submission(pred_ereg_mlm, pred_ereg_hlm, start_tm + "_VOT" + str(seed_num) + "_MDL" + str(use_models) +
"_UPSCALE" + "_RMSE" + str(int(rmse_mlm * 50 + rmse_hlm * 50)))
print("end...")
실행
C:\Users\wooha\anaconda3\envs\myPy39\python.exe C:\Users\wooha\PycharmProjects\newdrug\main_voting.py
vt_threshold : 0.05 , feature size = 259
RF MLM best parameter = {'max_depth': 14, 'max_features': 0.37415743749011454, 'min_samples_leaf': 3, 'min_samples_split': 12, 'n_estimators': 107, 'n_jobs': 20, 'random_state': 396}
RF HLM best parameter = {'max_depth': 21, 'max_features': 0.34523416743022384, 'min_samples_leaf': 1, 'min_samples_split': 2, 'n_estimators': 105, 'n_jobs': 20, 'random_state': 396}
20231027_192230 model_name = RF RMSE = 29.75 , MLM_RMSE = 28.520776327262347 , HLM_RMSE = 30.973266748471378
XGB MLM best parameter = {'colsample_bytree': 0.7433980076493739, 'eta': 0.06740788365834993, 'eval_metric': 'rmse', 'gamma': 9.963283702857769, 'max_depth': 8, 'min_child_weight': 14.0, 'n_jobs': 20, 'random_state': 396, 'reg_alpha': 160.0, 'reg_lambda': 0.19899962762618884, 'subsample': 0.33180588119453513}
XGB HLM best parameter = {'colsample_bytree': 0.5281299902771915, 'eta': 0.12839845945960932, 'eval_metric': 'rmse', 'gamma': 1.016004380934043, 'max_depth': 3, 'min_child_weight': 4.0, 'n_jobs': 20, 'random_state': 396, 'reg_alpha': 152.0, 'reg_lambda': 0.36503937884472615, 'subsample': 0.7186782533208999}
20231027_192230 model_name = XGB RMSE = 29.98 , MLM_RMSE = 28.630107348008778 , HLM_RMSE = 31.32853957363349
LIGHTGBM MLM best parameter = {'learning_rate': 0.077120387849475, 'max_depth': 19, 'n_jobs': 20, 'num_leaves': 56, 'random_state': 396, 'reg_lambda': 0.646351009953103, 'subsample': 0.8338203837704764, 'tree_learner': 'serial', 'verbose': -1}
LIGHTGBM HLM best parameter = {'learning_rate': 0.09387850045479688, 'max_depth': 5, 'n_jobs': 20, 'num_leaves': 122, 'random_state': 396, 'reg_lambda': 0.751478463234045, 'subsample': 0.4362515696508934, 'tree_learner': 'data', 'verbose': -1}
20231027_192230 model_name = LIGHTGBM RMSE = 29.84 , MLM_RMSE = 28.56743728907483 , HLM_RMSE = 31.104874976810123
HGB MLM best parameter = {'learning_rate': 0.032080015532927504, 'max_depth': 6, 'max_iter': 400, 'max_leaf_nodes': 21, 'min_samples_leaf': 10, 'random_state': 396}
HGB HLM best parameter = {'learning_rate': 0.0833558500354453, 'max_depth': 15, 'max_iter': 100, 'max_leaf_nodes': 28, 'min_samples_leaf': 7, 'random_state': 396}
20231027_192230 model_name = HGB RMSE = 29.89 , MLM_RMSE = 28.618333903201886 , HLM_RMSE = 31.152722863794523
EXTRATREE MLM best parameter = {'max_depth': 87, 'max_features': 0.21986248412535497, 'min_samples_leaf': 2, 'min_samples_split': 5, 'min_weight_fraction_leaf': 1.8110669525399563e-07, 'n_estimators': 208, 'n_jobs': 20, 'random_state': 396}
EXTRATREE HLM best parameter = {'max_depth': 77, 'max_features': 0.276062806333287, 'min_samples_leaf': 2, 'min_samples_split': 3, 'min_weight_fraction_leaf': 2.319771535625649e-06, 'n_estimators': 785, 'n_jobs': 20, 'random_state': 396}
20231027_192230 model_name = EXTRATREE RMSE = 29.93 , MLM_RMSE = 28.712442526972534 , HLM_RMSE = 31.156938374103053
Best models:
Rank 1 * : MLM = RF 28.520776327262347 HLM = RF 30.973266748471378
Rank 2 * : MLM = LIGHTGBM 28.56743728907483 HLM = LIGHTGBM 31.104874976810123
Rank 3 * : MLM = HGB 28.618333903201886 HLM = HGB 31.152722863794523
Rank 4 * : MLM = XGB 28.630107348008778 HLM = EXTRATREE 31.156938374103053
Rank 5 : MLM = EXTRATREE 28.712442526972534 HLM = XGB 31.32853957363349
20231027_192230 model_name = VOTING RMSE = 29.57 , MLM_RMSE = 28.110247749295542 , HLM_RMSE = 31.024087122925668
20231027_192230 model_name = VOTING UP-SCALE RMSE = 30.24 , MLM_RMSE = 29.14524476000838 , HLM_RMSE = 31.338726557606694
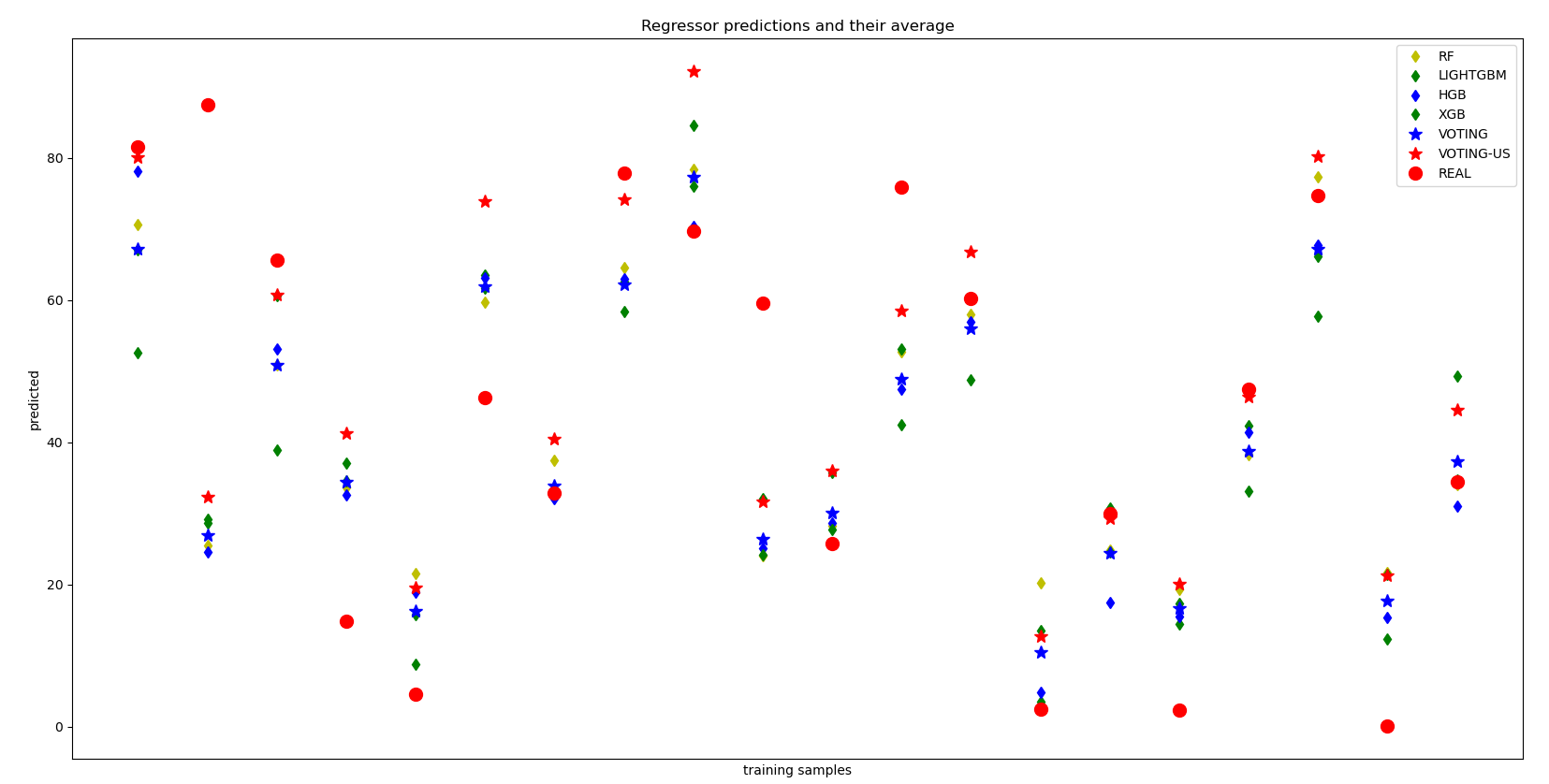
보팅 결과 비교
개별 모델의 예측 결과와 보팅한 결과를 실제와의 차이를 확인할 수 있습니다.
소스
'프로그램 > 파이썬' 카테고리의 다른 글
| 파이썬 키움증권 API 자동로그인 (0) | 2024.05.22 |
|---|---|
| 파이썬 32비트에 TA-Lib 설치하기 (0) | 2024.02.28 |
| 파이썬 패키지 설치시 OpenSSL 오류 (0) | 2024.02.26 |
| 파이썬 "Ignoring invalid distribution" 경고 조치법 (0) | 2023.08.01 |
| 파이썬 investing.com 크롤링(investpy) 우회 접근 (2) | 2023.07.12 |
댓글
공지사항
최근에 올라온 글
최근에 달린 댓글
- Total
- Today
- Yesterday
링크
TAG
- 메타에디터
- FX마진
- FX브로커
- fusiongrid
- RAPIDS
- cuda
- 전략프로그램
- maria
- zerotier
- 트레이딩시그널
- AI이미지생성
- 도커
- metaeditor
- 우분투
- GPU
- 신약개발 AI 경진대회
- pip check
- MySQL
- 메타트레이더
- copytrading
- Ignoring invalid distribution
- 스프링부트
- fx
- 파이썬
- 카피트레이딩
- ta-lib
- docker
- investy
- scraperlink
- 마진
| 일 | 월 | 화 | 수 | 목 | 금 | 토 |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| 8 | 9 | 10 | 11 | 12 | 13 | 14 |
| 15 | 16 | 17 | 18 | 19 | 20 | 21 |
| 22 | 23 | 24 | 25 | 26 | 27 | 28 |
| 29 | 30 | 31 |
글 보관함
